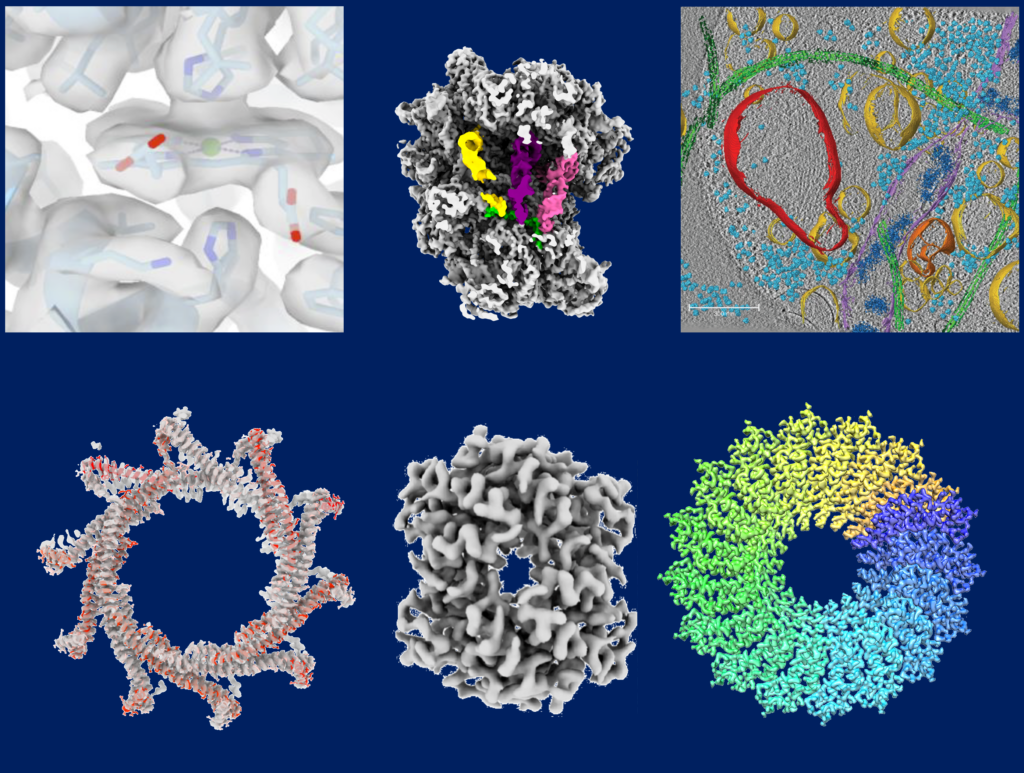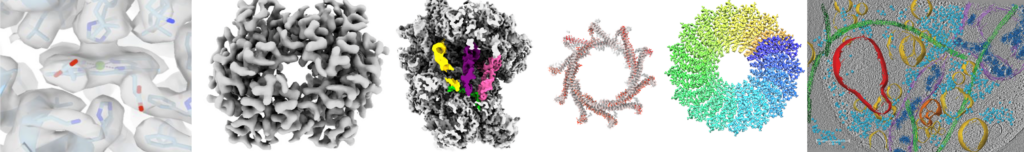
Cryo-electron microscopy (cryo-EM) is a revolutionizing technique used to determine the structures of biological molecules. The sample is frozen in a thin layer of vitreous ice and imaged in an electron microscope. The method allows imaging a large range of biological molecules such as small proteins in nano-meter scale as well as large macromolecular complexes like filamentous assemblies at highest resolution. Moreover, even entire viruses, prokaryotic and eukaryotic cells can be visualized in near-native frozen-hydrated state.
Selected Publications:
Schönnenbeck, P., Junglas, B. & Sachse, C. CryoVIA: An image analysis toolkit for the quantification of membrane structures from cryo-EM micrographs. Structure 33(4), 808–819.e4 (2025). https://doi.org/10.1016/j.str.2025.01.013
Junglas, B., Hudina, E., Schönnenbeck, P. Ritter, I., Heddier, A., Santiago-Schübel, B., F Huesgen, P., Schneider, D. & Sachse, C. Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies. Nat Struct Mol Biol 32, 23–34 (2025). https://doi.org/10.1038/s41594-024-01359-7
Junglas, B., Kartte, D., Kutzner, M., Hellmann, N., Ritter, I., Schneider, D. & Sachse, C. Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies. Nat. Struct. Mol. Biol. 32, 555–570 (2025). https://doi.org/10.1038/s41594-024-01399-z
Küçükoğlu B, Mohammed I, Guerrero-Ferreira RC, et al. Low-dose cryo-electron ptychography of proteins at sub-nanometer resolution. Nat Commun. 2024;15(1):8062. http://doi:10.1038/s41467-024-52403-5
Junglas B, Gewehr L, Mernberger L, et al. Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP. Cell Rep. 2024;43(9):114657. http://doi:10.1016/j.celrep.2024.114657
Knospe, C. V., Ortiz, J., Reiners, J., Kedrov, A., Gertzen, C. G. W., Smits, S. H. J. & Schmitt, L. Structural insights into the substrate binding mechanism of the class I dehydratase MadB. Commun. Biol. 8, 1032 (2025). https://doi.org/10.1038/s42003-025-08454-5